We can get the DNA sequence of the range of chromosome in human genome by using ucsc web tool.
http://genome.ucsc.edu/cgi-bin/das/hg19/dna?segment=chr8:126544471,126546471

We can get the DNA sequence of the range of chromosome in human genome by using ucsc web tool.
http://genome.ucsc.edu/cgi-bin/das/hg19/dna?segment=chr8:126544471,126546471

We can simply use the following formulae to extract the number from string in Excel file

=SUMPRODUCT(MID(0&E4, LARGE(INDEX(ISNUMBER(--MID(E4, ROW(INDIRECT("1:"&LEN(E4))), 1)) * ROW(INDIRECT("1:"&LEN(E4))), 0), ROW(INDIRECT("1:"&LEN(E4))))+1, 1) * 10^ROW(INDIRECT("1:"&LEN(E4)))/10)
Note: Tested in Excel 2010
We can use the tool picard to liftOver VCF file by batch
URL:
http://broadinstitute.github.io/picard/command-line-overview.html#LiftoverVcf

create dictionary (optional)
if you do not have the dict of reference fasta, you need to create first
java -jar /software/picard-tools-2.8.1/picard.jar CreateSequenceDictionary REFERENCE=/software/GATK_files/hg38/hg38.fa OUTPUT=/software/GATK_files/hg38/hg38.dict
download the chain file
Go the http://hgdownload.cse.ucsc.edu/downloads.html#human and download from liftOver hyperlink
liftOver
java -jar /software/picard-tools-2.8.1/picard.jar LiftoverVcf I=input.vcf O=lifted_over.vcf CHAIN=/software/liftOver/hg19ToHg38.over.chain.gz REJECT=rejected_variants.vcf R=/software/GATK_files/hg38/hg38.fa
Reference:
Chain format
URL: https://genome.ucsc.edu/goldenpath/help/chain.html
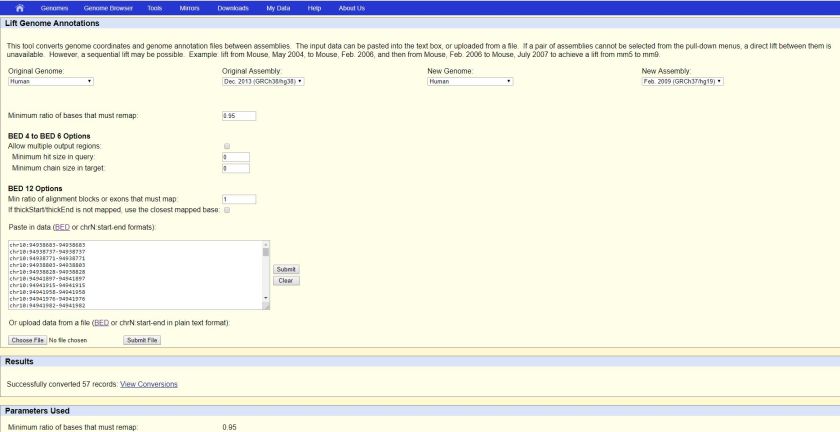
We can visit the web tool at https://genome.ucsc.edu/cgi-bin/hgLiftOver
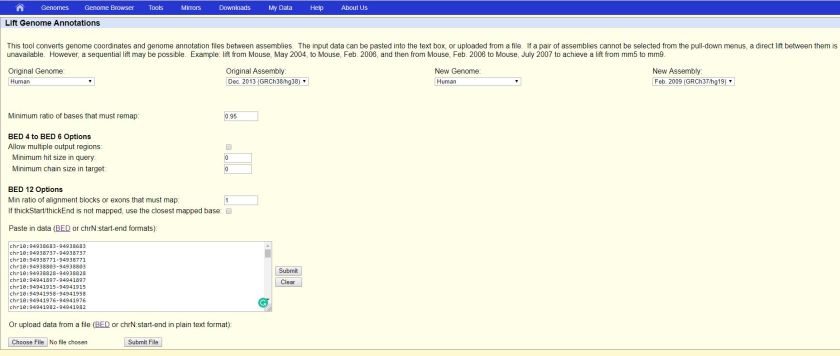
Select the Original Genome, Original Assembly, New Genome and New Assembly, say
Original Genome: Human
Original Assembly: Dec. 2013 (GRCh38/hg38)
New Genome: Human
New Assembly: Feb. 2009 (GRCh37/hg19)
Paste the data and click “Submit” button as the RHS of the text area
The result file (i.e. hglft_genome_260a_37bd80.bed) will be ready to download after some time
/software/samtools/1.8/bin/samtools bam2fq -N AGG0011.bam > AGG0011.fastq
cat AGG0011.fastq | grep ‘^@.*/1$’ -A 3 –no-group-separator > AGG0011_R1.fastq
cat AGG0011.fastq | grep ‘^@.*/2$’ -A 3 –no-group-separator > AGG0011_R2.fastq